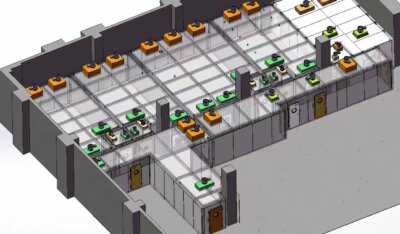
Antibiotic resistance is one of the most pressing global health challenges of our time. As bacteria evolve to withstand our most potent drugs, researchers are racing to understand these superbugs and develop new treatments. At the forefront of this battle are Biosafety Level 3 (BSL-3) laboratories, specialized facilities equipped to handle dangerous pathogens safely. These labs play a crucial role in investigating antibiotic-resistant bacteria and developing strategies to combat them.
The study of antibiotic-resistant bacteria in BSL-3 labs involves a complex interplay of advanced research techniques, stringent safety protocols, and cutting-edge technology. From isolating resistant strains to screening potential new drugs, these laboratories are vital in our ongoing fight against antimicrobial resistance. This article will delve into the world of BSL-3 labs, exploring their role in antibiotic resistance research, the challenges they face, and the groundbreaking work being done within their secure confines.
As we venture into this high-stakes realm of scientific research, we'll uncover the intricate processes involved in studying these resilient microbes. We'll examine the safety measures that protect researchers and the public, the advanced equipment used to analyze these dangerous pathogens, and the collaborative efforts driving progress in this critical field. By understanding the work conducted in BSL-3 labs, we can better appreciate the monumental task of addressing antibiotic resistance and the innovative approaches being developed to safeguard public health.
Biosafety Level 3 (BSL-3) laboratories are essential facilities in the fight against antibiotic-resistant bacteria, providing a secure environment for researchers to study and develop countermeasures against these dangerous pathogens.
What are BSL-3 laboratories, and why are they crucial for antibiotic resistance research?
BSL-3 laboratories are highly specialized research facilities designed to handle infectious agents that can cause serious or potentially lethal disease through inhalation. These labs are integral to the study of antibiotic-resistant bacteria due to their advanced containment features and rigorous safety protocols. They provide a secure environment where scientists can work with dangerous pathogens without risking exposure to themselves or the surrounding community.
In the context of antibiotic resistance research, BSL-3 labs offer a controlled setting for isolating, culturing, and analyzing resistant bacterial strains. These facilities are equipped with state-of-the-art technology and follow strict biosafety guidelines, allowing researchers to conduct experiments that would be too risky in lower-level laboratories.
The importance of BSL-3 labs in combating antibiotic resistance cannot be overstated. They enable scientists to study the mechanisms of resistance, test new antimicrobial compounds, and develop innovative strategies to overcome bacterial defenses. Without these specialized facilities, our ability to understand and address the growing threat of antibiotic-resistant superbugs would be severely limited.
BSL-3 laboratories provide an essential barrier between dangerous pathogens and the outside world, enabling critical research on antibiotic-resistant bacteria while ensuring the safety of researchers and the public.
| BSL-3 Lab Features | Purpose |
|---|---|
| Negative air pressure | Prevents contaminated air from escaping |
| HEPA filtration | Removes airborne particles and microorganisms |
| Restricted access | Limits entry to trained personnel only |
| Personal Protective Equipment | Protects researchers from exposure |
| Decontamination protocols | Ensures safe handling of materials and waste |
How do BSL-3 labs ensure safety when working with antibiotic-resistant bacteria?
Safety is paramount in BSL-3 laboratories, especially when dealing with antibiotic-resistant bacteria. These facilities employ multiple layers of protection to minimize the risk of exposure and prevent the release of dangerous pathogens into the environment. The design and operation of BSL-3 labs are governed by strict regulations and guidelines set forth by national and international health authorities.
One of the primary safety features of BSL-3 labs is the use of negative air pressure. This ensures that air flows into the laboratory rather than out, preventing contaminated air from escaping. All exhaust air is filtered through high-efficiency particulate air (HEPA) filters before being released, trapping any potentially harmful microorganisms.
Personal protective equipment (PPE) is another crucial aspect of BSL-3 safety. Researchers working with antibiotic-resistant bacteria must wear specialized protective clothing, including respirators, double gloves, and protective eyewear. Additionally, all work with live cultures is conducted within biological safety cabinets, which provide an extra layer of containment.
The multi-layered safety approach in BSL-3 labs, combining engineering controls, personal protective equipment, and strict protocols, creates a secure environment for studying antibiotic-resistant bacteria without compromising researcher or public safety.
Decontamination procedures are rigorously enforced in BSL-3 labs. All materials leaving the lab, including waste and equipment, must be sterilized or decontaminated. This often involves the use of autoclaves, chemical disinfectants, and other validated methods to ensure that no viable pathogens leave the facility.
| Safety Measure | Description |
|---|---|
| Airlock entry system | Prevents direct opening to external areas |
| Hands-free sinks and eyewash stations | Reduces contamination risk during emergencies |
| Sealed windows and surfaces | Facilitates decontamination and prevents leaks |
| Emergency protocols | Procedures for spills, exposures, and other incidents |
| Regular safety training | Ensures all personnel are up-to-date on safety practices |
What types of antibiotic-resistant bacteria are studied in BSL-3 labs?
BSL-3 laboratories are equipped to handle a wide range of antibiotic-resistant bacteria that pose significant threats to public health. These include both naturally occurring resistant strains and those that have acquired resistance through genetic mutations or horizontal gene transfer. The specific pathogens studied in BSL-3 labs are often classified as Risk Group 3 organisms, which can cause serious or potentially lethal human disease but for which preventive or therapeutic interventions may be available.
Some of the most commonly studied antibiotic-resistant bacteria in BSL-3 labs include:
- Multidrug-resistant Mycobacterium tuberculosis (MDR-TB)
- Methicillin-resistant Staphylococcus aureus (MRSA)
- Vancomycin-resistant Enterococci (VRE)
- Carbapenem-resistant Enterobacteriaceae (CRE)
- Extended-spectrum beta-lactamase (ESBL) producing organisms
These bacteria are of particular concern due to their resistance to multiple antibiotics, making infections caused by them difficult to treat. BSL-3 labs provide the necessary containment to study these organisms safely, allowing researchers to investigate their resistance mechanisms, virulence factors, and potential vulnerabilities.
BSL-3 laboratories serve as critical research hubs for studying the most dangerous antibiotic-resistant bacteria, enabling scientists to develop new strategies for diagnosis, treatment, and prevention of infections caused by these formidable pathogens.
In addition to these well-known resistant bacteria, BSL-3 labs also play a crucial role in identifying and characterizing emerging resistant strains. This proactive approach allows researchers to stay ahead of potential outbreaks and develop countermeasures before new resistant bacteria become widespread public health threats.
| Bacteria | Resistance Mechanism | Clinical Impact |
|---|---|---|
| MDR-TB | Multiple drug efflux pumps | Prolonged treatment, increased mortality |
| MRSA | Modified penicillin-binding protein | Limited treatment options, high healthcare costs |
| VRE | Altered cell wall precursors | Difficult to treat nosocomial infections |
| CRE | Carbapenemase production | Limited or no effective antibiotics available |
| ESBL | Extended-spectrum β-lactamase enzymes | Resistance to multiple β-lactam antibiotics |
What research techniques are employed in BSL-3 labs to study antibiotic resistance?
BSL-3 laboratories utilize a diverse array of research techniques to investigate antibiotic-resistant bacteria. These methods range from traditional microbiological approaches to cutting-edge molecular and genomic technologies. The combination of these techniques allows researchers to gain a comprehensive understanding of resistant bacteria and develop effective strategies to combat them.
One of the fundamental techniques used in BSL-3 labs is bacterial culturing and isolation. This involves growing bacteria on specialized media, often containing antibiotics to select for resistant strains. Researchers use these cultures to study the growth characteristics, morphology, and biochemical properties of resistant bacteria.
Advanced molecular techniques play a crucial role in antibiotic resistance research. Polymerase chain reaction (PCR) and DNA sequencing are used to identify specific resistance genes and mutations. Whole-genome sequencing has become an invaluable tool, allowing researchers to analyze the entire genetic makeup of resistant bacteria and track the spread of resistance genes across populations.
The integration of traditional microbiological methods with advanced molecular and genomic techniques in BSL-3 labs has revolutionized our understanding of antibiotic resistance, paving the way for targeted interventions and personalized treatment strategies.
Proteomics and metabolomics approaches are also employed to study the proteins and metabolic pathways involved in antibiotic resistance. These techniques provide insights into how resistant bacteria adapt to antibiotic stress and identify potential targets for new drugs.
High-throughput screening is another powerful tool used in BSL-3 labs. This technique allows researchers to quickly test thousands of compounds for antimicrobial activity against resistant bacteria. QUALIA has developed advanced screening platforms that accelerate the discovery of new antibiotics and other antimicrobial agents, offering hope in the fight against resistant pathogens.
| Research Technique | Application in Antibiotic Resistance Research |
|---|---|
| Bacterial culturing | Isolation and characterization of resistant strains |
| PCR and sequencing | Identification of resistance genes and mutations |
| Whole-genome sequencing | Analysis of genetic basis for resistance |
| Proteomics | Study of proteins involved in resistance mechanisms |
| Metabolomics | Investigation of metabolic adaptations in resistant bacteria |
| High-throughput screening | Discovery of new antimicrobial compounds |
How do BSL-3 labs contribute to the development of new antibiotics?
BSL-3 laboratories play a pivotal role in the development of new antibiotics to combat resistant bacteria. These facilities provide the necessary containment and resources to conduct essential research that bridges the gap between basic science and clinical applications. The work done in BSL-3 labs is crucial for identifying potential new drug candidates and understanding their effectiveness against resistant pathogens.
One of the primary contributions of BSL-3 labs to antibiotic development is in the screening of novel compounds. Using high-throughput screening technologies, researchers can rapidly test large libraries of chemical compounds against various antibiotic-resistant bacteria. This process helps identify molecules with promising antimicrobial activity that could be developed into new antibiotics.
BSL-3 labs also enable detailed studies of the mechanisms of action of potential new antibiotics. By observing how these compounds interact with resistant bacteria at the molecular level, researchers can optimize drug candidates for maximum effectiveness and minimize the potential for resistance development.
BSL-3 laboratories serve as crucial proving grounds for new antibiotic candidates, providing the secure environment necessary to evaluate their efficacy against the most dangerous resistant pathogens before advancing to clinical trials.
Another significant contribution of BSL-3 labs is in the development of combination therapies. By testing various combinations of existing and new antibiotics against resistant bacteria, researchers can identify synergistic effects that may overcome resistance mechanisms. This approach has led to the development of several successful combination treatments for multi-drug resistant infections.
The Antibiotic-resistant bacteria in BSL-3 labs facilities are equipped with advanced imaging and analytical tools that allow researchers to visualize the effects of antibiotics on bacterial cells in real-time. This capability provides valuable insights into drug efficacy and helps in the optimization of dosing strategies.
| Contribution | Impact on Antibiotic Development |
|---|---|
| Compound screening | Identification of potential new antibiotics |
| Mechanism studies | Understanding how new drugs overcome resistance |
| Combination therapy research | Development of more effective treatment regimens |
| Real-time imaging | Visualization of antibiotic effects on bacterial cells |
| Resistance evolution studies | Prediction and prevention of future resistance |
What challenges do researchers face when working with antibiotic-resistant bacteria in BSL-3 labs?
Working with antibiotic-resistant bacteria in BSL-3 laboratories presents numerous challenges that researchers must navigate to ensure both the safety of personnel and the integrity of their experiments. These challenges range from technical and logistical issues to the psychological stress of working with dangerous pathogens.
One of the primary challenges is maintaining the stringent safety protocols required in BSL-3 environments. The need for constant vigilance and adherence to safety procedures can be mentally and physically taxing for researchers. The use of personal protective equipment, while essential, can be cumbersome and may limit dexterity, making certain laboratory tasks more difficult to perform.
Another significant challenge is the complexity of working with highly resistant bacteria. These organisms often require specialized growth conditions and can be difficult to manipulate genetically. Researchers must continually adapt their methods to overcome these obstacles and obtain reliable results.
The challenges faced by researchers in BSL-3 labs, from maintaining rigorous safety standards to overcoming the technical hurdles of working with resistant bacteria, underscore the dedication and expertise required to advance our understanding of antibiotic resistance.
The potential for accidental exposure to antibiotic-resistant pathogens is a constant concern in BSL-3 labs. While safety measures are designed to minimize this risk, the consequences of exposure can be severe, given the limited treatment options for many resistant infections. This reality can create psychological stress for researchers working in these environments.
Lastly, the regulatory and ethical considerations surrounding work with antibiotic-resistant bacteria can pose challenges. Researchers must navigate complex approval processes and ensure their work complies with national and international guidelines on biosafety and biosecurity.
| Challenge | Impact on Research |
|---|---|
| Safety protocol adherence | Increased time and effort for lab procedures |
| Technical difficulties | Need for specialized techniques and equipment |
| Exposure risk | Psychological stress and potential health consequences |
| Regulatory compliance | Complex approval processes and documentation requirements |
| Ethical considerations | Balancing research needs with public safety concerns |
How are BSL-3 labs adapting to emerging threats in antibiotic resistance?
BSL-3 laboratories are at the forefront of addressing emerging threats in antibiotic resistance, constantly evolving their capabilities and approaches to stay ahead of this global health challenge. As new resistant strains emerge and spread, these facilities must adapt quickly to provide the research infrastructure needed to understand and combat these threats.
One key adaptation is the incorporation of advanced genomic technologies. Many BSL-3 labs now have on-site sequencing capabilities, allowing for rapid characterization of newly isolated resistant strains. This enables researchers to quickly identify resistance genes and track the spread of resistant bacteria in real-time, informing public health responses and guiding research priorities.
Another important adaptation is the development of more sophisticated in vitro models that better mimic human infections. These models, such as organoids and microfluidic devices, allow researchers to study how antibiotic-resistant bacteria behave in conditions that more closely resemble the human body, leading to more translatable research findings.
The ongoing adaptation of BSL-3 labs to emerging antibiotic resistance threats demonstrates the scientific community's commitment to staying at the cutting edge of research, ensuring we have the tools and knowledge to combat even the most formidable resistant pathogens.
BSL-3 labs are also increasingly adopting artificial intelligence and machine learning approaches to analyze the vast amounts of data generated in antibiotic resistance research. These tools can help identify patterns and predictions that might not be apparent through traditional analysis methods, potentially accelerating the discovery of new antibiotics and resistance mechanisms.
Collaboration has become a key strategy for BSL-3 labs in addressing emerging threats. Many facilities are now part of global networks that share data, resources, and expertise. This collaborative approach allows for a more coordinated and effective response to new resistant strains as they emerge around the world.
| Adaptation | Benefit |
|---|---|
| On-site genomic sequencing | Rapid characterization of new resistant strains |
| Advanced in vitro models | More accurate simulation of human infections |
| AI and machine learning | Enhanced data analysis and prediction capabilities |
| Global collaboration networks | Coordinated response to emerging threats |
| Biosecurity enhancements | Improved protection against potential misuse of research |
What role do BSL-3 labs play in global antibiotic resistance surveillance?
BSL-3 laboratories are integral to global antibiotic resistance surveillance efforts, serving as critical nodes in the worldwide network monitoring the emergence and spread of resistant bacteria. These facilities provide the secure environment and technical expertise necessary to identify, characterize, and track antibiotic-resistant pathogens of concern.
One of the primary roles of BSL-3 labs in surveillance is as reference centers for confirming and characterizing resistant isolates. When healthcare facilities or regional laboratories encounter bacteria with unusual resistance patterns, they often send samples to BSL-3 labs for detailed analysis. This process ensures accurate identification and reporting of resistant strains, which is crucial for informing public health responses.
BSL-3 labs also contribute to surveillance by conducting regular screening of environmental and clinical samples to detect emerging resistance trends. This proactive approach helps identify new resistant strains before they become widespread, allowing for early intervention and containment strategies.
BSL-3 laboratories function as sentinels in the global fight against antibiotic resistance, providing early warning of emerging threats and generating the data necessary to guide public health policy and research priorities.
Many BSL-3 facilities participate in national and international surveillance networks, sharing data on resistant isolates and contributing to global databases. This collaborative effort enables the tracking of resistance patterns across geographic regions and helps identify global trends in antibiotic resistance.
Additionally, BSL-3 labs play a crucial role in validating and improving diagnostic methods for detecting antibiotic resistance. By working with the most challenging resistant strains, these labs can develop and refine tests that are then distributed to clinical laboratories worldwide, enhancing global capacity for resistance detection.
| Surveillance Activity | Impact on Global Antibiotic Resistance Monitoring |
|---|---|
| Reference testing | Accurate characterization of resistant isolates |
| Environmental screening | Early detection of emerging resistant strains |
| Data sharing | Contribution to global resistance databases |
| Diagnostic validation | Improvement of resistance detection methods |
| Trend analysis | Identification of global patterns in antibiotic resistance |
In conclusion, BSL-3 laboratories are indispensable in the ongoing battle against antibiotic-resistant bacteria. These specialized facilities provide the secure environment necessary to study dangerous pathogens, develop new treatments, and monitor the global spread of resistance. The work conducted in BSL-3 labs spans from basic research into resistance mechanisms to the testing of novel antibiotics and the validation of diagnostic tools.
The challenges faced by researchers in these labs are significant, ranging from maintaining stringent safety protocols to overcoming the technical difficulties of working with highly resistant organisms. However, the dedication of scientists and the continuous adaptation of BSL-3 facilities to emerging threats ensure that we remain at the forefront of antibiotic resistance research.
As we look to the future, the role of BSL-3 labs in combating antibiotic resistance will only grow in importance. These facilities will continue to serve as crucial hubs for innovation, collaboration, and surveillance in the global effort to address one of the most pressing public health challenges of our time. By supporting and expanding BSL-3 research capabilities, we invest in our ability to understand, prevent, and overcome the threat of antibiotic-resistant bacteria, safeguarding public health for generations to come.
External Resources
BSL3 Biomedicum-SciLifeLab Collaborative Platform – Describes a BSL-3 laboratory at Karolinska Institutet equipped to handle various risk-group 3 pathogens, including antibiotic-resistant bacteria.
RBL NIAID BSL-3 Priority Pathogens – Lists priority pathogens that can be handled in a BSL-3 laboratory, including various antibiotic-resistant bacteria.
Laboratory Risks for Brucellosis – CDC – Provides guidelines for handling Brucella species, which are often antibiotic-resistant, in a laboratory setting.
New BSL-3 lab to advance research on pathogens – Rockefeller – Discusses the establishment of a new BSL-3 laboratory at Rockefeller University for studying highly infectious and antibiotic-resistant pathogens.
Biosafety Levels (BSL) – CDC – Provides comprehensive information on the standards and protocols for BSL-3 labs, crucial for handling antibiotic-resistant bacteria.
High-Throughput Screening for Antimicrobial Resistance – Discusses high-throughput screening methods applicable to identifying compounds effective against antibiotic-resistant bacteria in BSL-3 labs.
- Guidelines for Biosafety Laboratory Competence – WHO – Provides guidelines for biosafety laboratory competence, including standards for handling antibiotic-resistant bacteria in BSL-3 settings.
Related Contents:
- Effluent Decontamination Systems: Combating Antibiotic Resistance
- BSL-3 Lab Pathogens: Risk Group 3 Agents Guide
- BSL-4 Viral Research: Protocols for High-Risk Agents
- Emergency Response: Mobile BSL Labs in Action
- BSL-4 Biodefense: Cutting-Edge Research Programs
- Compact BSL-4 Labs: Innovative Design Trends
- BSL-3 vs BSL-4: Key Differences in Lab Safety Levels
- BSL-3+ Labs: Enhanced Features for Biosafety
- Unveiling Mobile BSL-3 and BSL-4 Lab Differences